Part A
Argument 1
The most important aspect we should consider while designing relational database schema is the normalization standard. Normalization is a process of organizing the data in database to avoid data redundancy, insertion anomaly, update anomaly & deletion anomaly [1].
For 1NF, the database is quite good since all the attributes are atomic for usage. However, it fails to meet the requirements of 2NF. The definition of 2NF is the following [2]. A relation is in 2NF if it is in 1NF and no non-prime attribute is dependent on any proper subset of any candidate key of the relation. A non-prime attribute of a relation is an attribute that is not a part of any candidate key of the relation.
There are three violations of 2NF in original design. We know the candidate key of the table is [PID, CHR, POS, RSID]. But there are three fields only partially depend on the candidate keys. Gender only depends on PID, pvalue only depends on RSID,age only depends on the PID. Those structure violates the principles of 2NF, which may cause serious problem. To be more specific, data stored is redundant with large storage overhead, some value are stored in multiple places, which means it is impossible to do single-point update.
Argument 2
When looking at the original design, it is clear that many unnecessary redundancies are introduced. One of the most obvious features is that since GWAS_Diabetes_M_20150905 and GWAS_Diabetes_F_20150905 are already introduced, there is no need to store the table called GWAS_Diabetes_ALL_20150905, because Male table schema together with Female table schema together represent all the information that could appear in ALL schemas. And if we throw ALL schemas away, there is no performance overhead since no join is introduced. So having ALL schemas doubles the storage space we need and bring nothing about performance. Even worse, someone may just insert experiment only in ALL schemas or only in Male schema, which might cause inconsistency of data, having bad effect on future possible data analysis. The same problem also exists for the ALL nationality schema.
Actually if these kinds of ALL tables are really needed in the actual situation, we can write it into a procedure or virtual view so that for the end user, there is no difference.
Argument 3
Another problem is that we store all data together; there is no distinct level for protection. But we know patient sensitive information is more important than the pvalue, according to the principles in information security, those information is supposed to store separately so that different level of access rules can be made to protect patient privacy.
Argument 4
The end users may be interested in different part of data, some of them want to do statistical analysis on trial results, some of them want to view patient information, and some of them want to see the experiments done through a time frame. But since all of the information is stored together and no abstraction is made, all users should work with all information part of which is irrelevant with them. This would cause overhead on maintenance and work.
Part C
1 Design Decision
The assumption for my design of data warehouse is very general, which most of data warehouse should obey. The data warehouse is supposed to be subject-oriented, integrated, time-variant, nonvolatile collection of data. In our context, subject is the result of experiments, since the main usage of the database is to manage thousands of experiments. It should have nonvolatile constraints which mean the data is seldom changed once it is inserted and is mainly used for querying and reading.
The goal of the system is to manage the experiment data. The main fact table should focus on Experiment, along experiment there are four dimensions, date dimension, group dimension, nationality dimension and Trait dimension. We think it is not a good idea to encode Meta data information into the table name, making that information had to retrieve all SQL statements. So we set up four tables to store those meta information. This kind of design can enable analysis in those dimensions like slice and dice operations, which can make flexible data visualization.
Also as we analyzed before, the storage row of each experiment table conflicts with the normalization standard, which will bring a lot of trouble. Here we decompose the table into three pieces, as Patient Dim Trail Dim and SNP Dim.
SNP Dim only stores information about the SNP piece such as pvalue, patient dim store information of each patient like age while Trail dimension store data about different trail items in the experiments. The fully designed structure is not only obeying 2NF, but also conforms to 3NF purpose. This kind of design reduces a lot of storage overhead and potential inconsistency. Of course join is introduced, but since the foreign key can be physically indexed, it would not cause large problems.
As for public dataset, there is no single table aiming for that, because it would introduce so much join keys, which make the database more complicated, instead a proposed physical views are used and updated periodically, which reduce storage overhead as well as better privacy protection.
It is an obvious advantage over our solution against the original one. First of all, our design schema is more easy to extend, for the original one, every field is fixed, if you want to add more information or new columns, you have to add null values for the previous data. But in our design, the new data can be added and associated through foreign key reference. Secondly, the data warehouse design with fact table and dimension tables enable view the data in different aspects and granularity more easily. You can analysis the experiment in the dimension of date, gender or nationality as well as with different granularities such as month year or day. If you want to do this for the original design, very complicated SQL statements have to be written. Thirdly, since the data base schema is normalized into 2NF or higher, it reduces a lot of redundancy which is introduced by the original design. Lastly, the sensitive information of patient is isolated so that it is easier to provide privacy protection. However, there are still some disadvantages of our system, since the schema is normalized, the join is introduced when querying and analyzing, which slow down the query process, but if we can use physical index and batch operation efficiently, the bad effect can be avoided to some extent. So to summarize, although joining is introduced, our design solution still outperforms the original database design in various ways.
What’s more, when we look at the difference between highly abstract data model vs. an explicit data model, we can see highly abstract data model like ER diagram has the following advantages, first of all it is simple and clear, which can used to check and analyze the requirement of the clients and since it is dependent of DBMS, many details can be ignored at this stage, which helps us focus more on structure design than detailed implementations. However, explicit data model like tables in database is more important when we want to solve specific questions such as performance, speed, availability security and so on. Since explicit data model is associated with one DBMS, it is easy to check and verify design choice.
Finally, the patient sensitive data is isolated into a single table, which provides better data protection possibilities. And since all data is partitioned into different table using real world logic, users of different tasks can only focus on their own data without knowing the complexity of other tables, which also give potential possibility for multitasking and parallelism. The clear structure also facilities the database admin to manage each table and provide public information.
2. Hadoop
When we want to choose either Hadoop or relational database as our data backend service, we have to compare their difference in terms of usage. A database is a means of storing information in such a way that information can be retrieved from it. In simplest terms, a relational database is one that presents information in tables with rows and columns. A table is referred to as a relation in the sense that it is a collection of objects of the same type (rows). Data in a table can be related according to common keys or concepts, and the ability to retrieve related data from a table is the basis for the term relational database [3]. Hadoop is a Java-based programming framework that supports the processing and storage of extremely large data sets in a distributed computing environment [4]. There are three key technologies, HDFS, which is used for storing PB-size files in distribution environment; Hbase, which is column storage for large databases, MapReduce, which offer parallel computing solution for data analysis. According to the book Hadoop: The Definitive Guide [5], the difference of Hadoop and relational database can be shown in the following graph
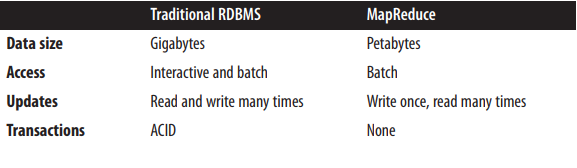
Traditional database usually store data at the level of Gigabytes while Hadoop is used of PB-level size data storage. Batch access is the only way for Hadoop access, while traditional database is more interactive, write can be done many times. ACID properties are well supported by RDBMS while Hadoop don’t provide mechanism for such properties.
For the GenomeDB application, data integrity is very important since the analysis of biomedical data should always be concise and consistent. The referential integrity is less important than data integrity but it could not be ignored, because sometimes we want to track each trial record to different experiments. But just for data analysis for traits, it isn’t as important as data integrity.
For referential integrity, relational database can do better. It can ensure there is no break foreigner key by delete triggers or some other constraints. However, Hadoop support Hbase column store, which has totally no referential integrity [6]. It is a tradeoff for performance. For data integrity, both of them have robust support, but I would say Hadoop is better, since geographical replication is implemented for the data and checksum algorithms are used to avoid corrupted cases.
Since data integrity is more important, we might prefer Hadoop as the backend platform. There is also another reason for doing this. GenomeDB is a typical example of embarrassingly parallel, since the trial of experiment is inserted once and mainly for reading. Each experiment is done for a specific trait, date, gender and population. So the analysis across the four dimensions is totally irrelevant to each other, no communication is needed and they can be fully parallelized.
What’s more Hadoop’s MapReduce programming architecture provides comprehensive and simple abstraction for implementing highly parallelized computation. If you use relational database, all kinds of parallelism have to be written manually by programmers. The analysis and query often involves large volume and statistical operation is often needed, which means Hadoop gives lots of performance gain.
Finally, the relational database usually fits on one single machine, if the volume begin large over years, it is complicated to scale out since join need to be done across machines. But for Hadoop, it is pretty easy to add more commodity hardware, the HDFS system will take care of them and user can only work in high abstraction level. For the end user, he doesn’t need to care all complexities behind Hadoop, so there is no so much maintenance overhead in the future.
[1]"What are the disadvantages of data redundancy?", Reference, 2017. [Online]. Available: https://www.reference.com/technology/disadvantages-data-redundancy-9a30353bc8edf864. [Accessed: 20- Oct- 2017].
[2] Codd, E.F. "Further Normalization of the Data Base Relational Model." (Presented at Courant Computer Science Symposia Series 6, "Data Base Systems," New York City, May 24th-25th, 1971.) IBM Research Report RJ909 (August 31st, 1971). Republished in Randall J. Rustin (ed.), Data Base Systems: Courant Computer Science Symposia Series 6. Prentice-Hall, 1972.
[3]"A Relational Database Overview (The Java™ Tutorials > JDBC(TM) Database Access > JDBC Introduction)", Docs.oracle.com, 2017. [Online]. Available: https://docs.oracle.com/javase/tutorial/jdbc/overview/database.html. [Accessed: 20- Oct- 2017].
[4]"Introduction to Hadoop – DZone Big Data", dzone.com, 2017. [Online]. Available: https://dzone.com/articles/introduction-to-hadoop-1. [Accessed: 21- Oct- 2017].
[5]Tom White., Hadoop: The Definitive Guide, Second Edition. [Place of publication not identified]: O'Reilly Media, Inc., 2010.
————————————————————————————————————————————————
